Introduction
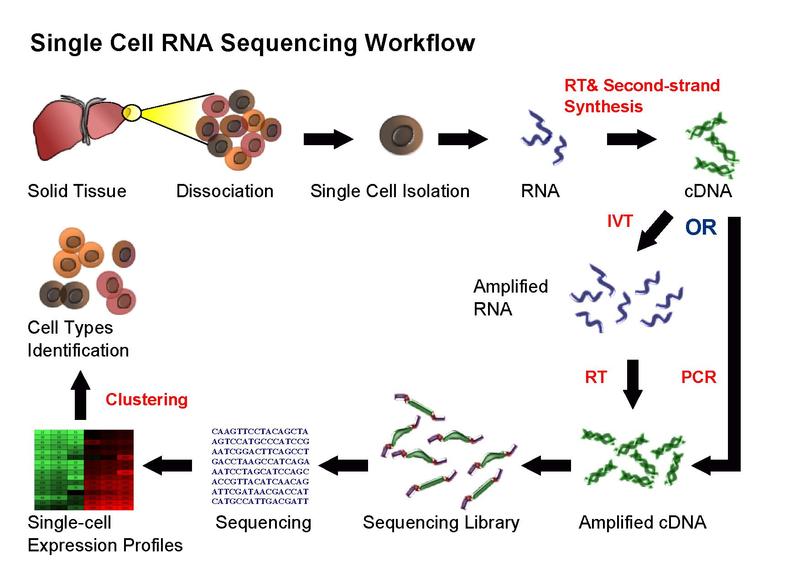
Single-cell RNA sequencing (scRNA-seq) provides the expression profile of individual cells. Through genes clustering analyses, rare cell types within a cell population can be identified, thereby making characterization of the subpopulation structure of a heterogeneous cell population become available. Single-cell RNA sequencing on numerous single cells can identify uncommon RNAs such as the RNA with low copy number, which may exert important functions in the cells, and also reveal the copy-number distribution of the whole mRNA population in individual cells.
Despite the advances in sequencing technologies, it is still unattainable to sequence RNA directly from single cells. Thus, in the current scRNA-seq protocols, RNA still needs to be converted to cDNA for sequencing. Principally, the current scRNA-seq methods contain the following steps: isolation of single-cell and RNA, reverse transcription (RT), amplification, library generation and sequencing.
| No | Headline | Click | Author | Date |
| 1 | Plant and Animal Whole Genome Re-Sequencing | 990 | Leading Biology | 2018-01-26 |
| 2 | Whole Exome Sequencing | 1019 | Leading Biology | 2018-01-26 |
| 3 | Whole Transcriptome Shotgun Sequencing | 1508 | Leading Biology | 2018-01-26 |
| 4 | smallRNA/microRNA/circRNA/LncRNA Sequencing | 979 | Leading Biology | 2018-01-26 |
| 5 | Bacterial Genome Sequencing | 1062 | Leading Biology | 2018-01-26 |
| 6 | Targeted Gene Sequencing | 1129 | Leading Biology | 2018-01-26 |