Introduction
Next-generation sequencing technologies made rapid assembly of draft microbial genomes possible, and sequencing the entire microbial genome is important for generating accurate reference genomes, for microbial identification, and other comparative genomic studies. Unlike traditional methods, NGS-based microbial genome sequencing doesn’t rely on labor-intensive cloning steps, saving time and simplifying the workflow. NGS can identify low-frequency variants and genome rearrangements that may be missed or are too expensive to identify using other methods.
The high-throughput sequencing is one of the NGS methods that we could use for the bacterial genome sequencing, including:
(1). identification of target DNA sequences and antigens to rapidly develop diagnostic tools;
(2). precise strain identification for epidemiological typing and pathogen monitoring during outbreaks;
(3). investigation of strain properties, such as the presence of antibiotic resistance or virulence factors.
The procedures are as follows:
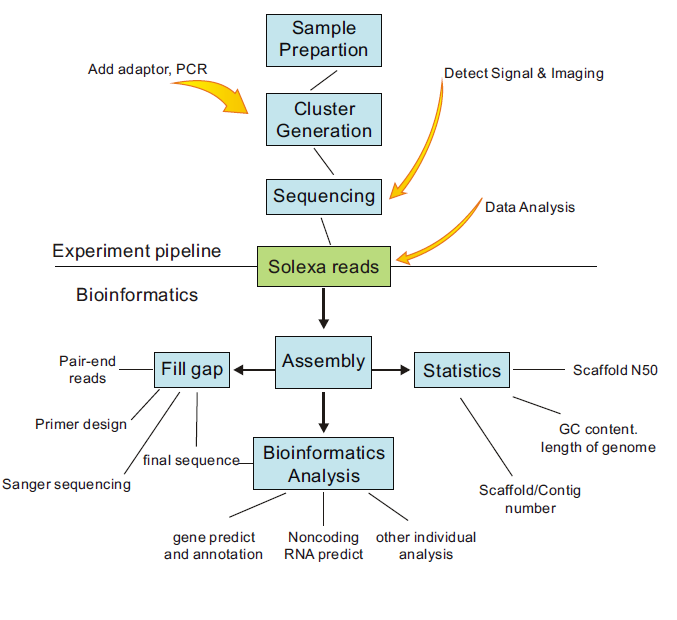
| No | Headline | Click | Author | Date |
| 1 | Plant and Animal Whole Genome Re-Sequencing | 989 | Leading Biology | 2018-01-26 |
| 2 | Whole Exome Sequencing | 1019 | Leading Biology | 2018-01-26 |
| 3 | Whole Transcriptome Shotgun Sequencing | 1508 | Leading Biology | 2018-01-26 |
| 4 | smallRNA/microRNA/circRNA/LncRNA Sequencing | 979 | Leading Biology | 2018-01-26 |
| 5 | Bacterial Genome Sequencing | 1061 | Leading Biology | 2018-01-26 |
| 6 | Targeted Gene Sequencing | 1129 | Leading Biology | 2018-01-26 |