> Custom Services > Sequencing and Chip Technology > Whole Transcriptome Shotgun Sequencing Introduction
Whole Transcriptome Shotgun Sequencing (WTSS), also called RNA sequencing, uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment in time.
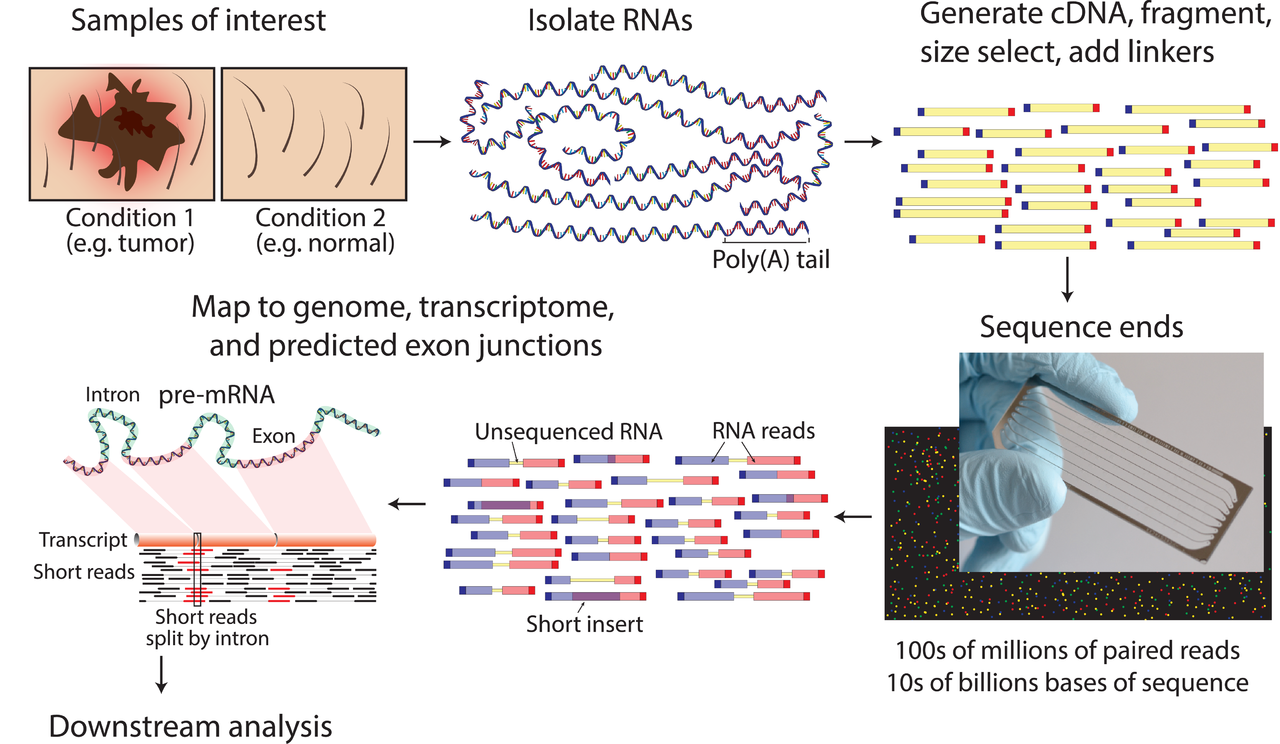
WTSS including a wide variety of applications from simple mRNA profiling to discovery and analysis of the entire transcriptome, including both coding mRNA and non-coding RNA (e.g., miRNA, small RNAs, linc RNAs). These applications are extremely popular for NGS platforms as they uncover information that may be missed by array-based platforms, as no prior knowledge of the transcript sequence is needed. Also, as it is sequencing-based, it is well suited for specialty applications such as RNA editing and allele-specific expression. While there are a variety of RNA-Seq applications and protocols, most follow the basic strategy of isolating RNA (such as with poly dT to pull down mRNA), converting it to DNA and then adding adaptor sequences to generate a library suitable for sequencing.
The WTSS procedures are as follows:
| No | Headline | Click | Author | Date |
| 1 | Plant and Animal Whole Genome Re-Sequencing | 990 | Leading Biology | 2018-01-26 |
| 2 | Whole Exome Sequencing | 1019 | Leading Biology | 2018-01-26 |
| 3 | Whole Transcriptome Shotgun Sequencing | 1508 | Leading Biology | 2018-01-26 |
| 4 | smallRNA/microRNA/circRNA/LncRNA Sequencing | 980 | Leading Biology | 2018-01-26 |
| 5 | Bacterial Genome Sequencing | 1062 | Leading Biology | 2018-01-26 |
| 6 | Targeted Gene Sequencing | 1129 | Leading Biology | 2018-01-26 |