> Custom Services > Sequencing and Chip Technology > smallRNA/microRNA/circRNA/LncRNA SequencingsmallRNA/microRNA Sequencing
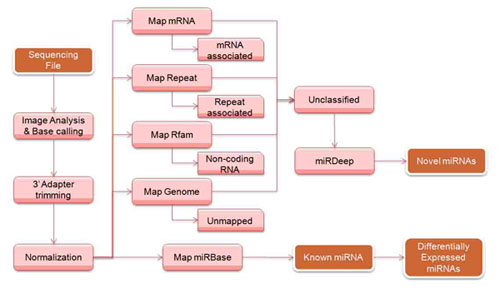
Small non-coding RNAs act in gene silencing and post-transcriptional regulation of gene expression. Small RNA sequencing is a technique to isolate and sequence small RNA species, such as microRNAs (miRNAs).
With small RNA-Seq, you can discover novel miRNAs and other small non-coding RNAs, and examine the differential expression of all small RNAs in any sample. You can characterize variations such as isomiRs with single-base resolution, as well as analyze any small RNA or miRNA without prior sequence or secondary structure information.
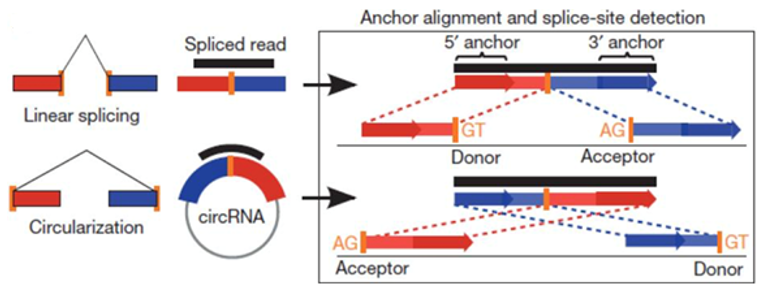
circRNA sequencing
Circular RNAs (CircRNAs) are a recent novel class of abundant, stable and ubiquitous RNAs addition to the growing list of types of non-coding RNA molecules, they can arise from both exons (exonic circRNA) and introns (intronic circRNA) and serve as miRNA or RNA-binding protein ‘sponges’, sequestering miRNAs and preventing their interactions with target mRNAs, as a result, controlling transcriptional events.
LncRNA Sequencing
Long non-coding RNAs (lncRNAs) are emerging as an important class of regulatory transcripts that are implicated in a variety of biological functions. RNA-sequencing, along with other NGS-based approaches, enables the study of lncRNAs on a genome-wide scale, at maximal resolution, and across multiple conditions.
LncRNAs are generally classified into three groups based on their genomic regions:
(1). long intergenic ncRNAs (lincRNAs)
(2). intronic ncRNAs (incRNAs)
(3). natural antisense transcripts (NATs), which are transcribed from the complementary DNA strand of their associated genes.
LncRNA sequence information can be acquired with single-base resolution via the high-throughput sequencing by constructing the strand-specific library for the removal of rRNA, and identify the known lncRNA and predict the new one and its target region by leveraging of the powerful bioinformatics analysis platform. The identification of long non-coding RNAs (lncRNAs) relies on the detection of transcription from genomic regions that are not annotated as protein coding, such as regions that are devoid of open reading frames. Alternatively, looking for lncRNAs and mRNAs whose expression is correlated, can perhaps indicate co-regulation or related functions.
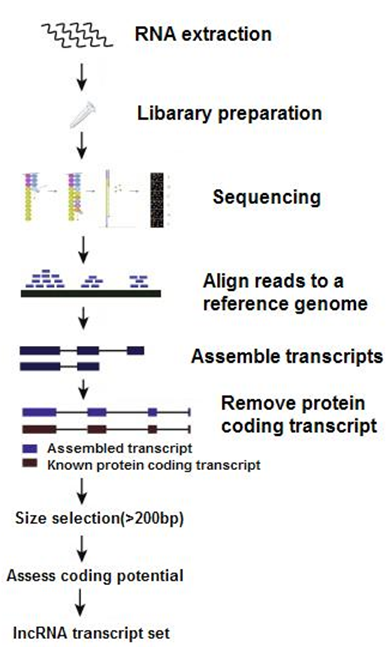
The lncRNA sequencing procedures are like below:
| No | Headline | Click | Author | Date |
| 1 | Plant and Animal Whole Genome Re-Sequencing | 990 | Leading Biology | 2018-01-26 |
| 2 | Whole Exome Sequencing | 1019 | Leading Biology | 2018-01-26 |
| 3 | Whole Transcriptome Shotgun Sequencing | 1508 | Leading Biology | 2018-01-26 |
| 4 | smallRNA/microRNA/circRNA/LncRNA Sequencing | 979 | Leading Biology | 2018-01-26 |
| 5 | Bacterial Genome Sequencing | 1062 | Leading Biology | 2018-01-26 |
| 6 | Targeted Gene Sequencing | 1129 | Leading Biology | 2018-01-26 |