> Custom Services > Immunology Services > Genome Editing: Which Should I Choose, TALEN or CRISPR?Introduction
Genomic editing - the ability to make specific changes to targeted genomic sites - is critical for biological and medical researchers (Bogdanove and Voytas, 2011; van der Voystet et al., 2013). Two genomic editing techniques have recently emerged using bacterial systems for plant pathogenesis or adaptive immunity: TALEN (transcription activator-like nuclease) and CRISPR (clustering, periodic intervals, and short Parindro repeating separately). TALEN and CRISPR both use endoenzymes to initiate double-stranded fractures (DSBs) in almost any genome target sequence and are used in many applications, including gene knock-off, genetically modified knock-on, gene markers, and gene defect correction. While both technologies are popular, the decision to choose one technology over another is not always clear, and Gene Copoeia customers occasionally ask us for advice. In this technical note, we compare the advantages and disadvantages of TALEN and CRISPR in order to provide customers with sufficient information to choose which technology to use to order reagents from us.
Genome editing
Genome editing starts with efficient DSB generation in the target DNA (Figure 1). DSB sits through homologous reorganization (HR) or through non-homologous end-to-end connection (NHEJ) without a homologous repair template. NHEJ causes small insertions or deletions because broken ends are pieced together. This tendency towards the indel generation is being used as a convenient way to knock out genes. TALEN consists of a pair of DNA binding proteins and CRISPR (Cas9 nuclease) and target-specific mono-conductor RNA (sgRNA) that can be edited by HR or NHEJ.
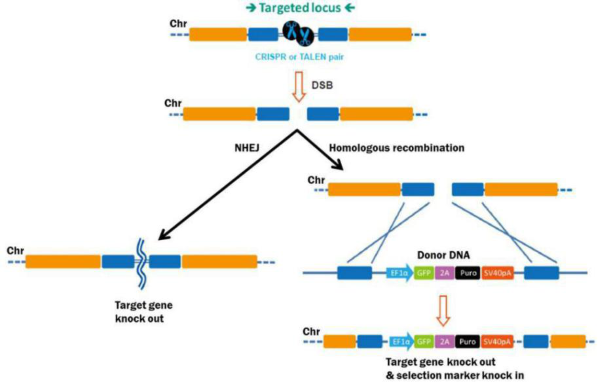
Figure 1. Pathways for repair of DSBs induced by genome editing tools. Left: Non-homologous end joining. Right: Homologous recombination in the presence of a donor template.
Comparisons between TALEN and CRISPR
There are four main concerns to think about when trying to decide between TALEN and CRISPR:
1. Specificity
Both TALEN or CRISPR provides high target site specificity, enabling researchers to make precise genetic alterations. CRISPR achieves this specificity by sgRNA, an artificial fusion of two naturally occurring short RNA (Jinek, et al., 2012). SgRNA directs S.pyogenes Cas9 nucleases to 20 nucleotide targets on chromosomes, which must be followed by N-G-G trinucleotides, called protocellular adhesion Motif, or PAM (Figure 2). SgRNA is cross-breeded with a chain opposite the PAM site, Cas9 nuclease cutting DNA. Many recent papers have shown that CRISPR can lead to the formation of DSBs at very high frequencies at the intended target site.
However, sgRNA can tolerate up to five mismatches (non-Watson-Crickkey-point pairing) and unwanted target sites (Fu et al., 2013), and CRISPR has been shown to be physically associated with many non-target sites in the genome (Kuscu et al., 2014). In addition, some studies have reported very high levels of Neder formation in unintended locations (Fu et al., 2013), but others have shown that initial estimates of deviation-target mutagenesis may be exaggerated (Li et al., 2013; Yang et al., 2013; Wang et al.,2014).
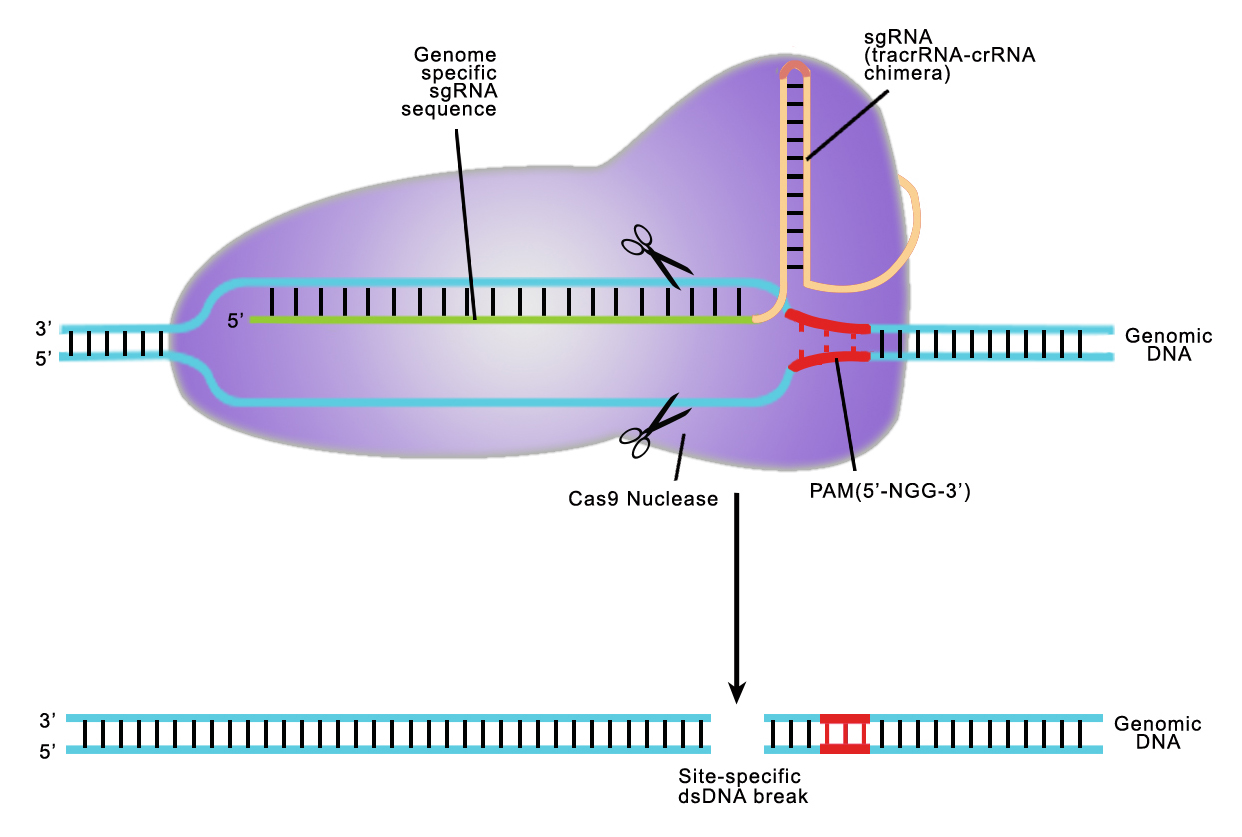
Figure 2. CRISPR-Cas9 target recognition.
Recent developments have significantly improved the specificity of CRISPR. One strategy uses a pair of single-chain fractures ("nickase") mutants that require targeting two sgRNAs (Mali et al., 2013) on the opposite strands on both sides of the target site; Ran et al., 2013). Individual nicknames are still recruited outside the target, but the nicks are smaller than the DSB's desperation, so pairings of nicknames can significantly reduce the deviation from the target. In addition, the reduction of sgRNA to 17 nucleotides also reduced off-target (Fu et al., 2014). Finally, the fusion between inactive Cas9 and FokI endasse (RNA-guided FokI nuclease, or "RFNs"), which requires that off-target mutations be reduced even in pairs of nickasas and truncated sgRNA by shifting sgRNA pairs.
In contrast, non-target activities do not seem to be a problem for TALEN. Typically, TALEN is built from 18 duplicate s34 amino acids. Repeat in amino acids 12 and 13, "repeated variable two components", or RVD changes. DNA binding code mediated by RVD (Figure 3) provides DNA binding specificity. TALEN pairs must be bound to the other side of the target site, separated by the "serats" of 14-20 nucleotides (Figure 4). This offset design is necessary because FokI requires duptication. As a result, this extremely long (about 36bp) DNA binding site is expected to be rarely found in the genome, if any. There are some degradations in the RVD-DNA binding code (Bogdanove and Voytas, 2011), but there is no evidence of mismatched tolerances or off-target activity in TALEN. For example, in a recent study, IPS cell lines were edited using highly active TALEN, and mutagenic activity was not detected at other genomic sites in the same as the target site (Park et al., 2014).
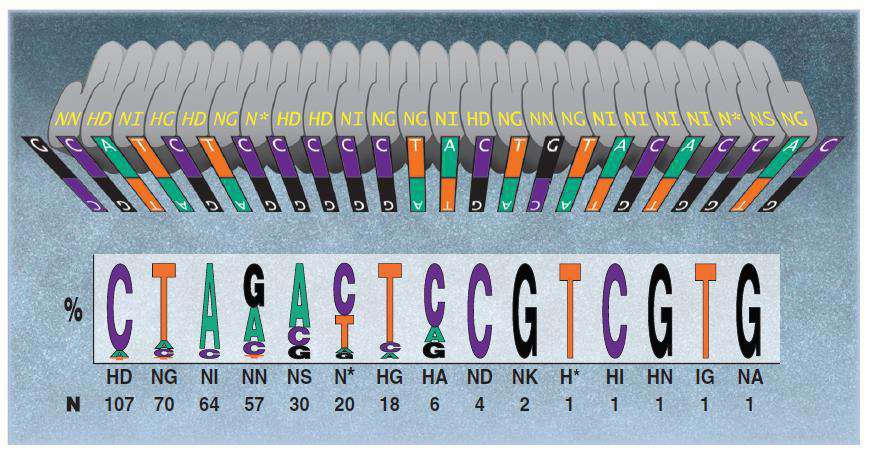
Figure 3. DNA binding code for TALENs. From Bogdanove & Voytas (2011).
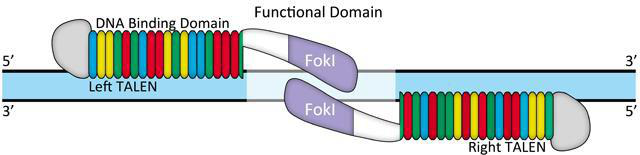
Figure 4. Typical TALEN design.
2. Target site selection
As mentioned above, the CRISPR sgRNA target must be immediately following the N-G-G-bit. It is often not difficult to find GG sites for a knockout, but this constraint can sometimes cause problems for other applications. In addition, dual notch enzymes and RFN strategies have additional target design limitations. For both, the two guide RNA must be oriented towards the PAM site distance from each other ("tail-to-tail" direction; Ran et al., 2013; Shen et al., 2014; Cai et al., 2014). In addition, the spacing between the tails of sgRNA is a key factor affecting CRISPR design. The paired nickass works best at offset distances between -8 and 30 bp. A 13-18 bp spacing is required, with the best pitch at 16-17 nucleotides. In conjunction with the requirements of the PAM site, paired CRISPR policies are sometimes difficult to implement in some applications.
On the other hand, while the TALEN design requires offset binding proteins with defined spacing, no other design constraints have been described. Therefore, in principle, the TALEN pair should be more free and flexible in target site selection than CRISPR can locate any bit in the genome.
3. Efficiency
CRISPR is popular in part because it modifies chromosome targets at high frequencies. More than 70% of indel formation rates have been reported. Truncated sgRNAs, pairs of nickases, and RFNs often reduce the efficiency of indel formation as expected.
TALEN is also able to modify chromosomes efficiently. In the example cited above (Park et al., 2014), a pair of TALEN pairs were measured in pieces to form 33%. This indel generation efficiency is comparable to that of all CRISPR versions. However, TALEN is sensitive to cytosine methylation, especially in CpG dinucleotides, a common and well-known MECHANISM of DNA silence. CpG methylation most commonly occurs in the promoter region, but it can also occur in protein-coded DNA. Some TALENs provide little or no mutagenic activity, and this sensitivity to methylation is a possible cause.
4. Ease of design and construction
CRISPR is simple to design and use. For each target, only 20 nucleotide genomic targets need to be programmed into the overall sgRNA. The plasmid structure is simple and straightforward. For editing experiments, sgRNA is expressed in conjunction with reusable Cas9 nucleases. The design and construction of sgRNA is the same for pairs of nickase and RFN methods, although in the latter case sgRNA must be selected and designed to function as a coordinating pair.
The reputation of the first generation of CRISPR is that the target design is almost always successful. Truncated sgRNA, paired nickases, or RFNs are not. Some designs are successful, while others are not, even if they are based on published data. In addition, CRISPR is not sensitive to methylation.
Although the TALEN structure involves redesigning new proteins for each target, the TALEN design has been simplified by the provision of repeated combination modules (reducing cloning). In addition, as mentioned earlier, TALEN is sensitive to cytosine methylation. One solution is to use N-RVD for 5 methylcytose (Figure 3), but it is important to know in advance that the target site has been methylated. Another workaround for this problem may be to avoid the CpG site. Of course, this will add another limitation, but if successful, TALEN will be more attractive as a genome editing tool.
Which should I choose, TALEN or CRISPR?
CRISPR is a relatively new technology, but TALEN still provides a valuable option due to relatively unconstrained target site requirements and high levels of specificity. When deciding whether to use TALEN or CRISPR, it is important to understand the differences between the two systems (Table 1). You should also consider the type of application you are trying. For example, if you're in a hurry to knock a gene out as part of a quick pilot study, the first generation of CRISPR is a great choice. Need fewer off-target mutagenesis in a relatively unconstrained design? Try TALEN.

Table 1. Comparisons between TALEN and CRISPR.
At Leading Biology, we offer products and services to CRISPR and TALEN. Our genome editing begins with expert design sits in non-viral and slow virus formats for TALEN and CRISPR vectors. In addition, we also provide a body-shaped design for HR-mediated applications. After the plasmid design, ready-made, sequence-validated plasmid DNA is built and delivered. In addition, we offer more complex genome editing services, such as target site validation, and the use of genome editing techniques for stable cell line and genetically modified mouse line generation. We also provide scientific advisory services where you can draw on our extensive knowledge and experience in this field. We will help you select the options that best suit your experiment and work with you to design a sound genome editing strategy by notifying you of the differences described in this document, among other things.
References
Bogdanove & Voytas (2011). TAL Effectors: Customizable proteins for DNA targeting. Science 333, 1843.
Fu, et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology 31, 822.
Fu, et al. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnology 32, 279.
Li, et al. (2013). Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 31, 681.
Jinek, et al. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816.
Kuscu, et al. (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nature Biotechnology.
Mali, et al. (2013). Cas9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnology 31, 833.
Park, et al. (2014). Targeted inversion and reversion of the blood coagulation factor 8 gene in human iPS cells using TALENs. Proc. Natl. Acad. Scie. U.S.A.
Ran, et al. (2013). Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 154, 1380.
Shen, et al. (2014). Efficient genome modification by CCRISPR-Cas9 nickase with minimal off-target effects. Nature Methods 11, 399.
Tsai, et al. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing van der Oost, et al. (2013). New Tool for Genome Surgery. Science 339, 768.
Wang, et al. (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80.
Yang, et al. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas- mediated genome
Related Services
Microorganisms Gene Modification Services
TALEN - Transcription activator-like effector nuclease
Contact Information
Please obtain a quote before ordering, and refer to the quote number when you place an order.
Orders are typically confirmed within 12 hours.
Have a Question? Email us info@leadingbiology.com
Order Products: Order Related Products
By Phone: 1-661-524(LBI)-0262 (USA)
| No | Headline | Click | Author | Date |
| 1 | Genome Editing: Which Should I Choose, TALEN or CRISPR? | 2269 | Leading Biology | 2020-01-03 |
| 2 | TALEN - Transcription activator-like effector nuclease | 3544 | Leading Biology | 2020-01-02 |
| 3 | Microorganisms Gene Modification Services | 1631 | Leading Biology | 2019-12-30 |
| 4 | CRISPR-CAS Technology | 2243 | Leading Biology | 2019-12-30 |
| 5 | Transgenic Plants Construction | 3064 | Leading Biology | 2019-12-30 |
| 6 | In Situ Hybridization | 3382 | Leading Biology | 2018-01-26 |